About us
Our research addresses the greatest challenges of the future: keeping societies healthy, fostering intact environments, promoting sustainable energy and developing well-functioning political and social systems. Seeking answers through interaction between a variety of disciplines, we develop concrete solutions for regional problems which can then be applied globally. We continually strive to open new paths with science and innovation, by responding to society’s complex questions with answers geared to the needs of people.
Facts
& Figures
See all Facts & Figures.
What we do
We develop concrete solutions for regional problems which can then be applied globally. We continually strive to open new paths with science and innovation, by responding to society’s complex questions with answers geared to the needs of people.
What makes us different
To undertake research is to challenge conventional ways of thinking and to transcend limitations. Our open enterprise culture and flat organizational structure allow this investigative spirit to develop in the best possible interdisciplinary way, and with curiosity, enthusiasm, courage and perseverance we highlight problems from all possible angles and thereby arrive at exceptional solutions.
Who we work with
We are linked to research networks all over the world with partners in more than 50 countries, on five continents. The research center collaborates with international organizations such as the Alpine and Carpathian Conventions, the United Nations Environment Programme (UNEP) and the European Space Agency (ESA) along with several other space agencies. It is also an official duty station of the United Nations and the only Italian headquarter of UNU (the United Nations University).Eurac Research furthermore cooperates with the Council of Europe and other European institutions.
Our history
Eurac Research was founded in 1992 as an association under private law with just twelve members of staff undertaking research in the areas of Language and Law, Minorities and Autonomous Regions as well as the Alpine Environment. The center gradually expanded its activities into new areas, attracted scientists from all over the world, and introduced new structures. Today, over 600 collaborators from almost 50 countries conduct their research at Eurac Research
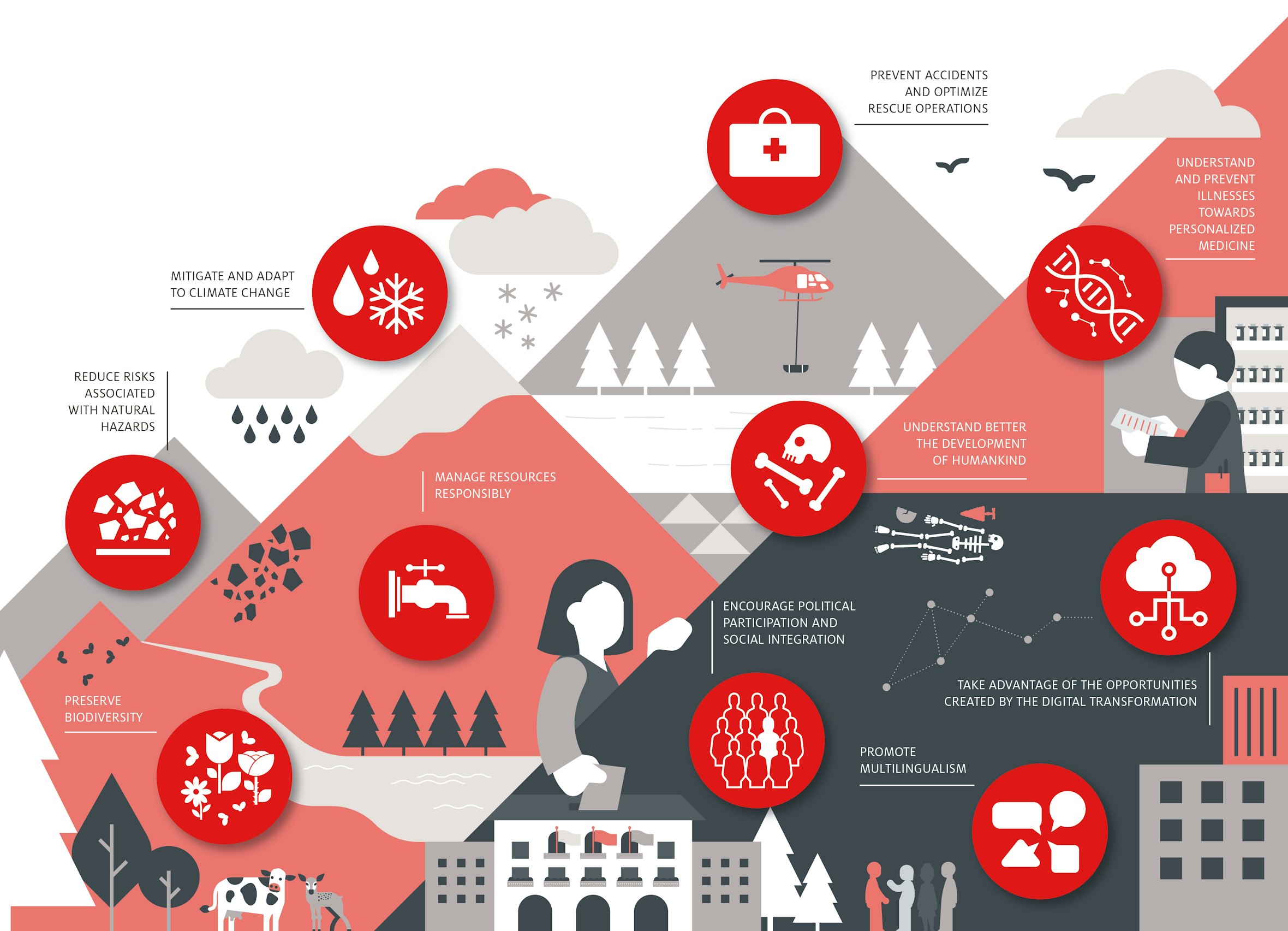
- Mitigate and adapt to climate change
- Manage resources responsibly
- Reduce risks associated with natural hazards
- Understand and prevent illnesses, towards personalised medicine
- Prevent accidents and optimise rescue operations
- Understand better the development of humankind
- Preserve biodiversity
- Encourage political participation and social integration
- Promote multilingualism
- Take advantage of the opportunities created by the digital transformation
Our goals
 © Eurac Research | Oscar Diodoro
© Eurac Research | Oscar Diodoro